Field Methods for the Identification of Emerging Viruses
PhD student: Anna Signe Fomsgaard
E-mail: annafomsgaard@gmail.com
THE PROJECT
Viruses are major contributors to novel emerging infectious diseases. Due to the versatile nature of viruses, human activities, contact with animals and environmental factors, the world is increasingly experiencing viral disease outbreaks. To recognise and respond quickly to such outbreaks, the first line of defence is knowing the variety of viruses circulating and being exchanged at the human-animal interface. However, many viral infections induce similar symptoms (syndromes), and some viruses can occur as co-infections and some are yet unknown. This makes it challenging to identify all or any specific virus. Furthermore, many of these pathogenic viruses are emerging in areas where expertise, plus high-technology facilities and infrastructure can be limited. This creates a window of time for the virus to spread uncontrollably. A fast identification system is pivotal for subsequent effective control or management plan. The development of metagenomic sequencing may reduce the challenge of identifying the disease-causing agent(s). Additionally, by enabling the transport of this technology out of the specialised laboratory and performing pathogen characterization at the point of incidence, viruses could be identified much earlier in an outbreak situation.
THE PURPOSE
The PhD project was part of the EU-funded TELE-Vir "Point-of-incidence toolbox for emerging virus threats” project. The purpose of the PhD project was to develop a fast field-deployable lab-in-a-bag for the characterization of viral agents at the initial site of occurrence. The work was based on technologies evaluated/developed to become field-deployable.
RESULTS
Study I. Responding to the international kit shortages during the onset of the COVID-19 pandemic, Study I presented a simple nucleic acid (NA) extraction method for SARS-CoV-2 diagnosis. The method involved incubating pharyngeal swab samples at 98⁰C for 5 minutes and cooling to +4⁰C. When used with the SensiFAST Probe No-ROX One-Step Kit and E-gene primers, this approach detected SARS-CoV-2 with 97.4% sensitivity, 100% specificity and 98.3% accuracy.
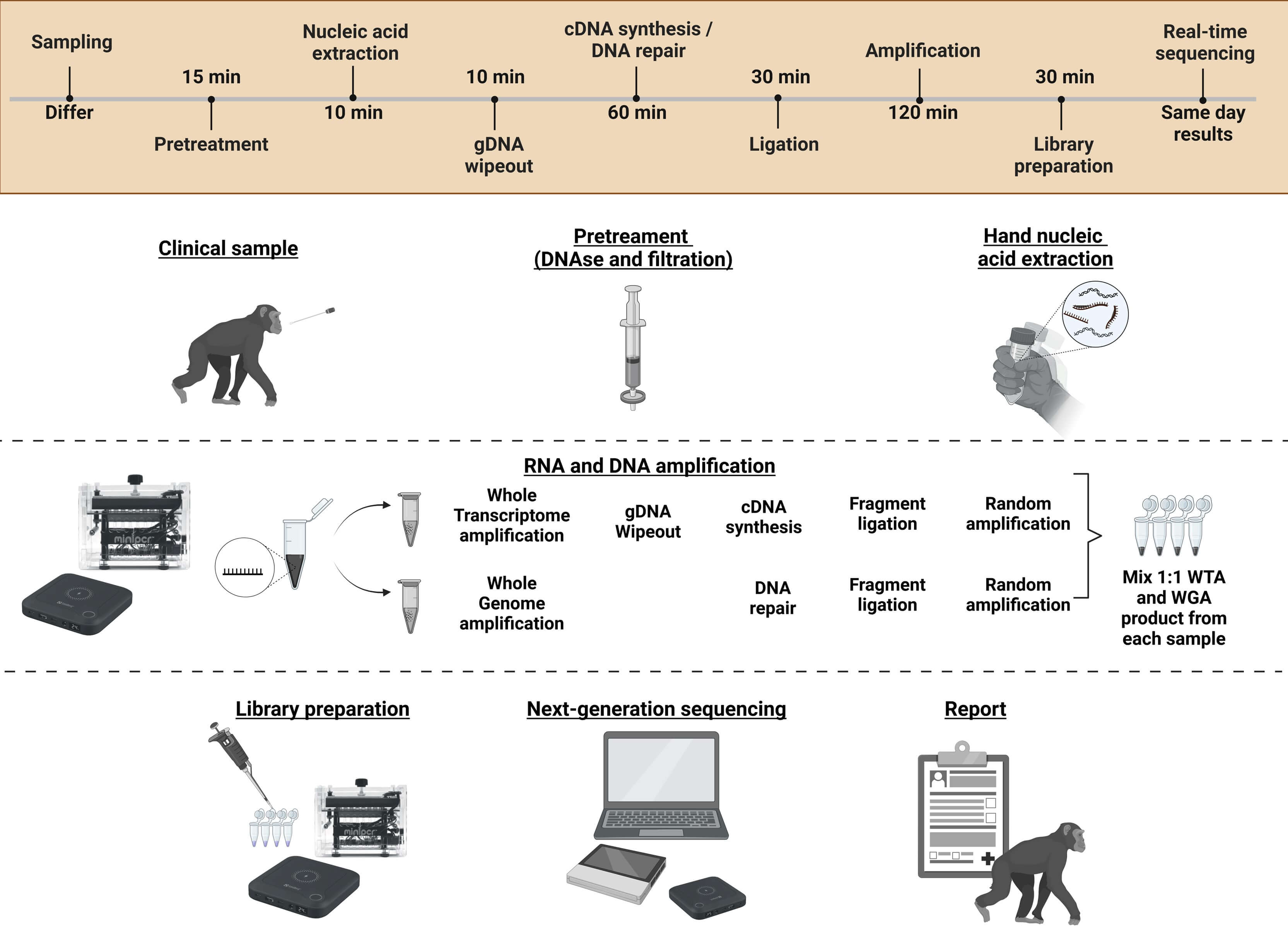
Study II. This study presented a metagenomic next-generation sequencing (mNGS) workflow for virus detection. The workflow combined handheld NA extraction methods, isothermal sequence-independent amplification, and real-time Nanopore sequencing. Key to virus detection success was a 15-minute table-top DNase I pretreatment, followed by syringe-based filtration, which increased viral read counts 500-fold compared to no pretreatment. This mNGS workflow demonstrated its proficiency in detecting SARS-CoV-2 and enabled lineage typing for samples with CT-values⪅30. It also identified the DNA virus, human papillomavirus (HPV), and detected co-infections with molluscum contagiosum virus, in cervical swabs.
Study III. The last study was dedicated to refining the mNGS workflow for field deployment. This study, conducted at Copenhagen Zoo over two months, processed samples from 37 vertebrate species across 11 types of sampling materials. Thirteen different vertebrate viruses from four different virus groups were identified and, for the first time, the suggested zoonotic Human Associated Gemykibivirus 2 was identified in the nasal swabs of a Linnaeus two-toed sloth (Choloepus didactylus). The workflow required only seven hours from sampling to sequencing, driven by portable power banks and hand-operated equipment.
THE FUTURE
Notwithstanding existing challenges, such as time, cost, differentiating closely related viruses and the overall complexity of data interpretation, the project presents methods wherein real-time, on-site virus detection is possible. This induces the hope of facilitating and improving public and veterinary health preparedness. The use of metagenomic sequencing technologies portends a future in which emerging viruses can be promptly identified and characterized. This applies not only in human populations but also in husbandry animals and vulnerable wildlife, thus hopefully mitigating the consequences of future viral disease outbreaks.
